The in situ hybridization technique is intended to detect a nucleotide sequence of interest by using its complementary nucleotide sequence, called probe. The complementarity, that is, the hybridization between the nucleotide sequence and the probe, is the base for the specificity of this technique. In this way, it can be studied when and where the expression of a particular gen is happening in a tissue, by detecting the messenger RNA, which tell us about the cell physiology. In situ hybridization can also be used for detecting the physical localization of a gen in a chromosome. In situ hybridization is not as widely used in histology labs as immunohistochemistry. However, it provides information about the physiology behavior of the cell that cannot be studied with other techniques. In situ hybridization may be performed on cell cultures, tissue sections or the entire animal, usually embryos. It is a technique very useful for studying embryo development, stem cell differentiation, gen manipulation or cell physiology.
Tissue
Sections, organs or embryos have to be treated before the probe is added for hybridization. As usual, samples need to be fixed. The common fixative is paraformaldehyde. It is better to perform hybridization on frozen sections, instead of paraffin sections, because the messenger RNA is better preserved. In addition, after fixation and cryoprotection, samples can be stored at -80 ºC for a long time, or samples can be fixed and immersed in methanol and stored at -20 ºC for months.
Single strands of RNA are easily degraded by RNAases, which are almost everywhere. Precautions have to be taken to prevent losing the RNA. It is mandatory to wear gloves because there is a huge amount of RNAase in our fingerprints, and lab-ware needs to be sterilized in an autoclave (not to remove bacteria but to inactivate RNAase). Other advisable step is a treatment of the tissues with proteases to improve the penetration and diffusion of the probe. Sometimes, it is convenient to block unspecific binding of the probe by acetylating with triethanolamine buffer and glacial acetic acid.
Probe
In situ hybridization is based on complementary nucleotides (A-T or A-U, and G-C). In the same way that immunohistochemistry uses antibodies, in situ hybridization uses a nucleotide sequence, or probe, which is complementary to the RNA sequence we want to detect. The probe is a single strand labeled with molecules that can be detected later. The size of the probe (the number of nucleotides that forms the sequence) usually ranges between 50 and 300 nucleotides, but larger probes can be used. Probes can be labeled with radioactive isotopes (isotopic hybridization) or conjugated with non-radioactive molecules (non-isotopic hybridization). Nowadays, non-isotopic hybridization is widely used and probes are frequently conjugated with biotin, fluorescent molecules, digoxigenin, bromodeoxyuridine, or other molecules.
Sometimes it is hard to get a proper probe, and this may be the reason hybridization is not so widely used in histology laboratories. The design (the sequence) and synthesis of the probe is not easy. Furthermore, the sequence has to be specific for the species we are working with. The same gene has no identical sequences in two species, hence the probe for one species does not probably work in the other. However, once a working probe is gotten, the hybridization technique is easy and reliable.
The first thing to get a working probe is to know the nucleotide sequence of the mRNA with want to detect. The sequence may be already available in some database in Internet and then a lot of effort is saved because nowadays customized nucleotide sequences can be purchased. Some companies are able to synthesize long DNA strands with customized sequences. DNA strands up to 1000 nucleotides are easy to get. Probes can be obtained by transcribing these sequences. Even, labeled probes can be ordered, saving time and money.
However, the sequence to make our probe is sometimes unknown. Thus, the first step is to figure out the messenger ARN (mRNA) sequence, which means that we need to clone the mRNA we want to detect. Cloning follows several steps. 1) Purification of the total RNA of the tissue and retrotranscription of the RNA into cDNA (cloned DNA), that is, the mRNA sequences are now in complementary DNA strands. 2) By using short sequences known as primers, which are specific for our sequence of interest, and the PCR (polymerase chain reaction) technique, multiple copies of our sequence are obtained. This process is known as amplification. 3) After purifying the amplified fragments, they are inserted in plasmids, and plasmids in bacteria. 4) Bacteriaare grown and yield many copies of the plasmid. Plasmids divide together with the bacteria DNA in each cell division. 5) Plasmids are then purified from bacteria colonies. The inserted fragment in the plasmid is sequenced to know if we have the sequence we are seeking. 6) The sequence is now transcribed, as in during S phase in normal cells, so that many RNAs are obtained. During the synthesis, some modified nucleotides are inserted in the probe. These nucleotides are conjugated with molecules, like biotin or digoxigenin that can be detected later by immunohistochemistry, or with fluorochromes that can be observed at fluorescent microscopy. These RNA labeled transcribed sequences are our probes.
Hybridization protocol
Once we have a proper probe, tissue is incubated in a solution containing the probe. Then, the hybridization between the probe and its complementary mRNA takes place. Those cells expressing the complementary mRNA are visualized by detecting the probe by immunohistochemistry (if the probe is labeled with digoxigenin (Figures 1, 2 and 3)) or other methods.
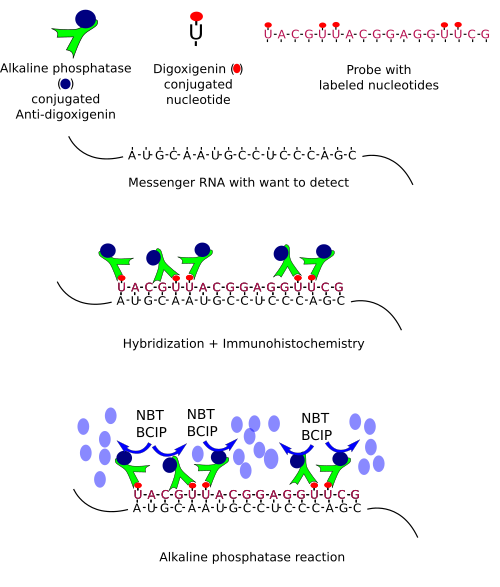
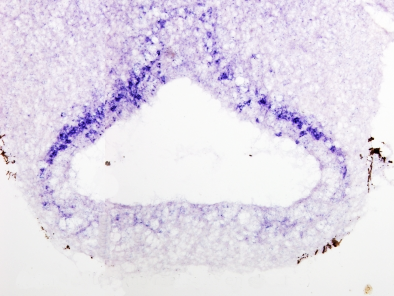
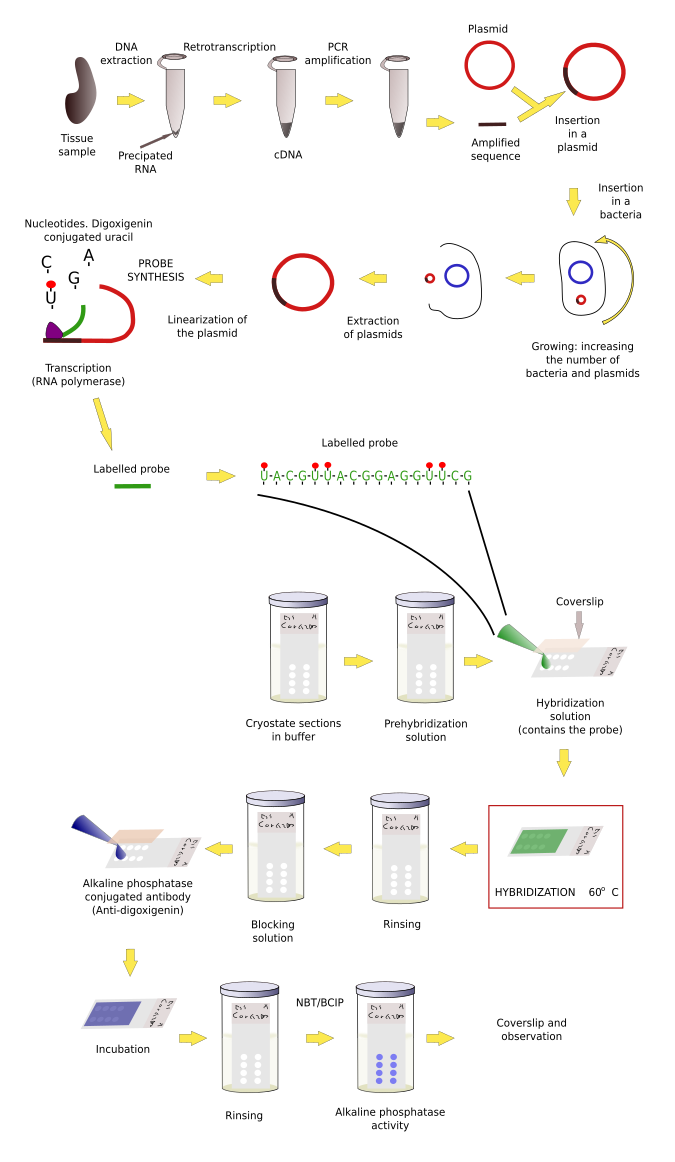
In situ hybridization can be combined with other techniques such as immunohistochemisty or general staining.
 Inmunohistochemistry
Inmunohistochemistry