1. ADN and histones
Chromatin is found in the nucleoplasm, surrounded by the nuclear envelope. Chromatin is DNA (deoxyribonucleic Acid) and associated molecules involved in DNA organization, primarily represented by histones. DNA is consists of 4 deoxyribonucleotides (abbreviated as nucleotides). Every nucleotide contains a nitrogenous base, a pentose and a phosphate group. The nitrogenous base is either a purine base: adenine (A) and guanine (G), or a pyrimidine base: thymine (T) and cytosine (C) (Figure 1). The pentose is a deoxyribose. Each nitrogenous base is linked to a pentose, resulting in a deoxyribonucleoside. Each deoxyribonucleoside is linked to a phosphate group through the pentose molecule, together forming a deoxyribonucleotide. In this way, DNA contains a chain of nucleotides linked by phosphate groups. This is a single chain, but DNA is made up of two strands, which are paired by the complementarity of the nucleotides. A-T and G-C, each nucleotide of each pair in different chains, establishes hydrogen bonds that keep the two DNA chains tightly joined. It is like rail tracks, where hydrogen bridges are the sleepers, and iron tracks are the phosphate-ribose strands. It is said that both chains join in an antiparallel manner, that is, the end of the rail track, because one of their ends is 3' in one chain and 5' in the other, whereas the other end is 5' in one chain and 3' in the other. Both chains are twisted in a double helix of about 2.5 nm in width.

Nucleotides are not exclusively found in the DNA. They also form other molecules with different functions. For example, ATP (adenosine triphosphate) is the major molecule for energy transfer, and cAMP (cyclic adenosine monophosphate) is a second messenger of many molecular signaling processes in the cell.
DNA is not naked in the nucleoplasm, but associated to proteins forming the chromatin . histones are DNA-associated proteins largely involved in chromatin organization. There are two types of histones: nucleosome histones (H2A, H2B, H3 and H4) and H1 histone. The four nucleosome histones, along with DNA, form the nucleosome. Nucleosome is the structural unit of chromatin organization. Other proteins are transiently associated to DNA, such as those involved in the DNA transcription (gene expression), DNA synthesis (replication), or chromatin condensation.
2. Heterochromatin y euchromatin
At transmission electron microscopy, the nucleoplasm shows clear and dark areas when observed during interphase (the non-mitotic part of the cell cycle) (Figure 2). Dark areas largely correspond to compacted chromatin, referred to as heterochromatin. Sometimes, a very dark and dense rounded area is observed in the nucleoplasm. It is the nucleolus. Nucleolus is made up of chromatin involved in the transcription of ribosomal RNA and ribosomal subunits assembling, as we will see in the next page. Heterochromatin is usually found close to the nuclear envelope and around the nuecleolus. Light areas contain more loosely arranged chromatin, known as euchromatin. Heterochromatina and euchromatin can also be observed at light microscopy (Figure 3).


Euchromatin usually includes DNA regions being activily transcribed, whereas heterochromatin largely consists of DNA regions not being transcribed or showing low transcription rate. Heterochromatin is classified into facultative heterochromatin, if it can alternate between euchromatin and heterochromatin, and constitutive heterochromatin, which is always condensed. Facultative heterochromatin is up to 10-20 % of the total heterochromatin. Although some genes have been found to be expressed in the heterochromatin, most are not. There is a higher level of chromatin compaction when chromosomes are formed, either in mitosis or in meiosis. During this process, both euchromatin and heterochromatin are packed into chromosomes.
3. Organization
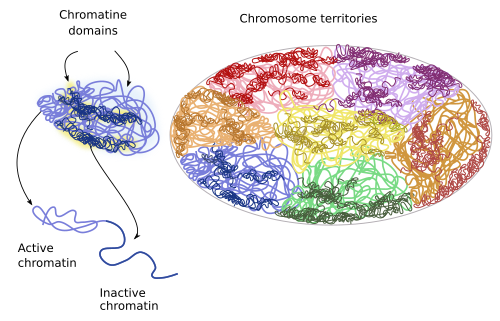
If we think that the nucleus in interphase is a mess of chromatin after chromosome decondensation, it is difficult to imagine how the cell is capable of managing chromatin to condensate, decondensate, and to regulate and express a specific set of genes without making a huge entanglement. Far from being a tangle, it is currently known that chromatin is not randomly distributed within the nucleus, and condensed and decondensed regions are specific portions of the chromatin. In 1980, it was observed that chromatin from a particular chromosome occupies a well-defined territory in the nucleoplasm. These territories are referred to as chromosomal territories. Each chromosome occupies its own territory, and each territory is in turn divided in the compartments A and B, which consists of many smaller regions and thousands of nucleotides. At a lower scale, there are topological associated domains, and finally chromatin loops of about hundreds of nucleotides. This organization is conserved in mammalian cells. The interactions of the chromatin with the nuclear lamina of the nuclear envelope help to segregate chromatin into territories and into other lower size compartments.
Chromosome territories. In interphase, each chromosome is found in a limited space within the nucleus, the chromosome territory (Figure 4). These territories are rather spherical, from 2 to 4 µm in diameter, and neighbor territories are only overlapped in the borders (in yeast, however, territories are not so well delimited). Many evidences support that the distribution of these territories in the nucleoplasm is not random. Although the distribution pattern may differ between cell types, differentiation stages, and along the cell cycle, it looks like that the spatial relation between chromatin territories is quite stable in the same cell type. More active territories, i.e. having more intense gene expression, are located toward the center of the nucleus, whereas the less active territories are closer to the nuclear envelope. In general, smaller chromosomes with many genes tend to be inner in the nucleus, while those large chromosomes bearing not too many genes are found peripherally. In the same territory, regions that are replicated during the first part of the S phase (early replication regions) are deeper than those replicated later in the S phase (late replication regions). Moreover, repressed genes have a preference for peripheral positions. It is remarkable that homologous chromosomes occupy separate regions within the nucleus.
A and B compartments. Chromosome territories are divided into the A and B compartments. The A compartment contains genes that are replicated early during the S phase of the cell cycle, and a high density o genes that are frequently expressed. The B compartment contains genes that are replicated late during the S phase and seldom expressed, and are located near the nuclear envelope. Some chromatin regions can switch between the two compartments. With improved resolution techniques, and regarding the particular modifications of histones, the compartment A has been subdivided into 2 additional regions, and B compartment into 3 regions.
Chromosome domains. They are small chromatin regions that usually interact with one another. For example, more than 2200 chromosome regions have being found in mouse stem cells. Curiously, these regions have not being found in plant cells. These domains are relatively stable in cell lineages, during cell differentiation, and even between different species. Because of this, chromosome domains have being claimed as the fundamental units of the genome and chromatin. There are repressed chromosome domains: polycomb chromatin, heterochromatin, and other less characterized types. The open chromosome domains, or open chromatin, are composed of actively expressed chromatin, and they can content regulatory sequences, promoters, expressed genes, and regions linked to insulating chromatin proteins. Despite this, the type of chromosome domain does not completely determine the behavior of a gene. Repressed chromatin does not usually interact with other chromatin domains, but open chromatin can do it, even with domains of other chromosome territories. The chromosome domains are the result of local interactions between chromatin loops.
Chromosome domains can be further subdivided. In addition, there are differences between chromatin loops, since some of them interact more frequently than others. It means that there are groups of chromatin loops that keep short distance between each other.
All these chromatin compartments are loss during the M phase of the cell cycle, when chromatin is compacted into chromosomes. It is not known how the chromatin compartments are formed again during chromosome decondensantion during the late phases of mitosis. In addition, during cell differentiation, it is possible to identify the usual compartments of a particular cell type. However, the interactions between compartments change.
-
Bibliography ↷
-
Bibliography
Cavalli G, Misteli T. 2012. Functional implications of genome topology. Nature structural and molecular biology. 20: 290-299.
Yu M, Ren B. 2017. The three-dimensional organization of mammalian genomes. Annual review of cell and developmental biology. 33: 265–89.
-
 Nuclear pore
Nuclear pore 